Role of post-translational modifications in regulation of tumor suppressor p53 function
Introduction
It has been widely accepted that p53 is an important tumor suppressor and TP53 is mutated in about 50% of all tumors. p53 germline mutation can lead to Li-Fraumeni syndrome, patients susceptible to various types of cancer (1). p53 knockout mice, with a higher incidence of cancer (mostly lymphoma), demonstrates the function of p53 as a tumor suppressor (2). In a wide range of human cancers, TP53 undergoes abnormally high frequency mutations. For example, TP53 mutations were identified in 50% to 60% of spontaneous human tumors such as the lungs, breast, bladder, colon, esophagus, stomach, liver, brain, bone, prostate, ovaries and lymphatic system tumors (3,4). p53 is also an important transcription factor that regulates the transcription of many downstream genes, such as p21 involved in cell cycle checkpoints, GADD45 involved in cell growth regulation and DNA damage repair, NOXA involved in the process of apoptosis (5,6). Therefore, p53 plays an essential role in regulating cell growth, DNA damage repair and apoptosis. The regulation of p53 transcriptional activity is important for its function as tumor suppressor. Research on p53 transcriptional activity is also essential for the treatment of p53 mutations -induced tumors. p53 can protect cells from the malignant development of tumors and play an important role in the process of aging, differentiation and fertility, as well as in neurodegenerative diseases, diabetes and myocardial infarction (7).
p53 is involved in many biological processes, and there are many questions required to be answered about the regulation mechanisms of p53 activity. How was a protein with so many important functions located at the right place at the right time? What mechanism is involved in regulating the various functions of p53? The regulation of p53 occurs at both the transcriptional level and the protein level. Under normal conditions for cell growth, p53 can be rapidly degraded by ubiquitin ligase MDM2. Genomic instability and a variety of other cellular stimuli lead to activation of p53 through many mediators such as ATM, p19ARF and CHK2 (8). Many effect proteins can then stabilize p53 protein and promote transactivation of downstream genes, including genes involved in DNA damage repair, cell cycle arrest, or programmed cell death. Protein post-translational modification is an important pathway for regulating protein activity and function. Similar to the N-terminal tail of histones, the C-terminal tail of p53 is a loose region containing many lysine sites that can undergo post-translational modifications. Modulation of p53 activity by post-translational modifications may be an explanation for p53 to participate in a large number of cellular functions. Different stimuli can lead to different post-translational modifications of p53, which can dynamically regulate its function.
There have been reported more than 50 loci involved in the post-translational modifications of p53 including phosphorylation, acetylation, methylation, ubiquitination, glycosylation, etc. (9,10). Many of these modifications occur in the response of stress, and are interdependent, then trigger a subsequent series of events (11). The development and application of antibodies that recognize site-specific modifications of p53 have greatly simplified the study of post-translational modifications of p53 proteins and have made rapid progress. In recent years, the application of mass spectrometry has promoted the identification of new post-translational modifications of p53.
Phosphorylation
Phosphorylation of p53 occurs mainly at serine and threonine residues of N-terminal and C-terminal. After the cells are stimulated, most of the phosphorylation occur at once. Also, there are some sites phosphorylated in normal situation and undergo dephosphorylation induced by DNA damage (12). p53 was identified as a phosphorylated protein shortly after its discovery, and the first identified phosphorylation sites were murine Ser312 and Ser389 (13). In 1992, Lees-Miller et al. found that DNA-activated protein kinase DNA-PK was able to phosphorylate Ser15 and Ser37 at the amino-terminal transactivation domain of p53 (14). After that, studies have reported that phosphorylation at Ser15 of p53 induced MDM2 dissociation with p53, leading to the stabilization of p53 protein (15). These are the earliest assumptions about the post-translational modifications of p53. A milestone discovery is that phosphorylation at Ser15 is essential for cell stress independent on DNA damage. In the glucose-induced response, Ser15 can be phosphorylated by the AMPK (AMP-activated protein kinase) pathway and mediates the metabolic cell cycle G1/S block (16). Ser6 and Ser9 were originally discovered as substrates for CK1δ and CK1ε, members of the protein kinase CK1 family, and were phosphorylated after treatment with multiple genotoxic and non-genotoxic agents. The phosphorylation at Ser6 and Ser9 is important for tumorigenesis and metastasis induced by TGF-β, activated Ras and mutant p53 (17). Phosphorylation at the two loci also plays crucial role in the mesodermal development of xenopus (18). Phosphorylation at Ser46 mediated by p38 MAPK, HIPK2, DYRK2, MDM4 (19) and possibly other kinase can exert apoptotic cell death, one possible mechanism is that phosphorylation at Ser46 induces amphiregulin expression and its target microRNA (20).The transactivation domain of p53 is capable of forming two domains with similar structure, TAD1 (1–40 residues) and TAD2 (41–83 residues). The TAD2 domain of p53 is capable of interacting with the p62 subfamily of the universal transcription factor TFIIH, which is important for the p53-initiated transcription events after loosening of the chromatin structure of the promoter region. Phosphorylation at Thr55 mediated by transcription factor TAF1 (TAFII250), as well as phosphorylation at Ser46 can trigger the interaction of TAD2 with pleckstrin homology (PH) region of p62 (21). Phosphorylation of p53 at position 392 can be induced by DNA damage and plays a role in activating the sequence-specific DNA binding capacity of p53. Phosphorylation may also stabilize the formation of p53 tetramers, which is important for its activity, so phosphorylation modification is essential for p53 to play suitable activity (22).
Acetylation
Acetylation at the lysine residue of p53 is induced in a variety of forms of cytotoxicity and non-cytotoxic stimuli, resulting in stabilization and activation of p53. Lysine residues of carboxyl terminal (K305, K370, K372, K373, K381, K382 and K386) and a DBD residue (K164) can be acetylated by CREB-binding protein (CBP) (KAT3A)/p300 (KAT3B), which can increase the sequence specific DNA binding capacity of p53 by inducing its conformational changes (23,24). K320 of p53 can be acetylated by another histone acetyltransferase p300/CBP-related factor PCAF (KAT2B) (25). The acetylation of p53 promotes the recruitment of transcriptional activators, such as CBP and PCAF complexes, in the promoter region of p53 downstream genes and activation of p53 downstream genes (26). Acetylation of p53 is important for its ability to inhibit the cell cycle progression of G2 phase, which is achieved by NF-Y-p53-dependent inhibition of G2 phase response gene (27,28). The effect of deletion of one or more sites may be compensated by other acetylation sites (29,30). Deacetylase HDAC1 could deacetylate most acetylation sites of p53 in vitro and cultured cells (31,32). Deacetylase SIRT1 is capable of interacting with p53 in the nucleus, specifically deacetylating the K382 acetylation of p53 (33). Depsipeptide, an inhibitor of HDAC, significantly induces acetylated p53 at K373/K382 binding to the regulatory region of p21 and increases the expression of p21 (23,34). Different stimulus signals can induce acetylation at K305 of p53 mediated by p300 in vivo and in vitro (35), which is important in regulating the transcriptional activity of p53 (35). K120, in the DNA binding domain of p53, can be acetylated by Tip60/hMOF (a MYST family HAT, independent of p300/CBP or PCAF) and the mutation of this site is of frequent recurrence in the process of tumor development (36). Acetylation at K120 induced by DNA damage significantly changes the effect of salt concentration on the specificity of its DNA binding capacity (37). p53 with K120 acetylation preferentially locates at the promoter region of the critical gene promoting apoptotic, rather than the promoter region of those genes involved in cell cycle arrest (38). Chronic myeloid leukemia (CML) is a disease that causes abnormal hematopoietic stem cell function due to the expression of BCR-ABL, which increased the acetylation at K317 of p53 and promoted the translocation of p53 to cytoplasm and activation of BAX after DNA damage. Acetylation at K320 (human p53)/K317 (mouse p53) plays an important role in the regulation of p53 shuttle between nucleus and cytoplasm and the p53-dependent BAX-mediated apoptosis following DNA damage (39). Recent research also revealed that in intestinal adenoma formation deacetylation of p53 was pivotal for the induction of autophagic flux (40).
In the absence of stimuli, p53 is at a low level of expression, mainly in the form of monomer. p53 protein has the most effective function in the form of tetramer because the DNA binding affinity of tetramer p53 is high (41). Oligomerization of p53 occurs prior to acetylation, and oligomerization could provide a docking site for acetyltransferase (42). Studies have shown that lysine acetylation modification of p53 C-terminal is of much higher efficiency in p53 tetramer than the dimmer and acetylation almost cannot occur on the p53 monomer. Acetylation at p53 C-terminal lysine residue also prevents the ubiquitination at the same lysine residue site induced by MDM2, further stabilizes the tetramer, enhances the DNA binding ability of p53 to specific sequence, and also promotes the recruitment of transcriptional activators (42).
The mechanism of p53 transactivation is variant on different promoters and acetylation modification plays an important role in the selective regulation of p53 function. The role of acetylation is dependent on the cellular environment.
Methylation
Methylation at lysine and arginine is also a reversible mechanism for the regulation of p53 function. K370, K372, K373 and K382 at the carboxy terminus of p53 can be methylated and the effect to enhance (43,44) or inhibit (45) the function of p53 depends on the modified site. Jansson et al. reported that three arginine residues R333, R335 and R337 of the p53 oligomerization domain (TET) can be methylated by a class II methyltransferase PRMT5 (46).
Lysine methylation
SET7/9 can methylate p53K372 in the nucleus after DNA damage, which enhances the overall stability of p53 binding to chromatin, increases recruitment of p53 in regulatory regions of p21 and other downstream genes, promotes transcriptional activation of p21 and other downstream genes (47). On the other side, Smyd2-mediated monomethylation at p53K370 inhibits the transactivation of p53, which inhibits p53-mediated cell cycle arrest and apoptosis (48). A demethylase KDM1, demethylating Lys370, prevents the binding of p53 coactivator 53BP1 (43). The 370 locus is close to the 372 locus, suggesting that interactions may occur between methylation at these lysines. Under physiological conditions, SET9 prevents Smyd2 from binding to p53 (48). The SET7/9-mediated methylation at K372 can activate p53 function by inhibiting Smyd2-mediated methylation at K370 after DNA damage. The role of smyd2-mediated methylation at K370 is to inhibit p53-mediated transcriptional activation; therefore, methylation at K370 after DNA damage repair is required to remove methylation at K372 so that the activity of p53 can be restored to the basal level. These results indicate that methylation at lysine is a dynamic post-translational modification in the complex regulation of p53 activity. However, p53K370 can also be bimethylated with the effect of positive regulation of p53 activity, one evidence is that after DNA damage p53 protein with K370me2 modification increased in the promoter region of p53 downstream gene. The demethylation process from activated bimethylation modification to inhibitory monomethylation is regulated by lysine-specific demethylase LSD1 (43). Researchers have identified another modification associated with p53 physiological function, monomethylation at p53K382 by SET8 in 2007, which reduces the transactivation of p53 to high response downstream genes (45). Different from the K382 monomethylation modification, the level of p53K382me2 increased after DNA damage and was recognized and bound by the tandem Tudor domain of 53BP1, which interacted with p53 by stabilizing the accumulation of p53 (49). In addition, studies have reported that G9a and Glp could mediate the bimethylation at p53K373, while the p53K373R mutants cannot be methylated. Different from the activation effect by K370me2 and K382me2 (mediated by interaction with 53BP1), p53K373me2 modification is a non-activated signal (50).
Arginine methylation
Arginine methylation can also modulate the activity of p53, which is an important regulatory mechanism in p53 response. When DNA is damaged, Strap is able to recruit PRMT5 to p53 and promote methylation of p53. The absence of PRMT5 changes the specificity of p53 binding in the promoter region and triggers apoptosis dependent on p53. Methylation by PRMT5 also has influence on the activity of p53 oligomerization (46). Under physiological conditions, the arginine methylation sites of endogenous p53 by PRMT5 were identified as Arg333, Arg335 and Arg337. Arg335 and Arg337 can be bimethylated by PRMT5 while Arg333 is monomethylated (51). Arginine methylation modulates the promoter binding specificity of p53, and PRMT5 siRNA reduces the protein level of p21, an important downstream protein of p53 involved in the regulation of cell cycle arrest. Whereas the proteins encoded by downstream genes of p53 involved in apoptosis, such as PUMA, NOXA, AIP1 and APAF1 (52) are little affected or unaffected. Importantly, mutations of Arg333 and Arg337 exist naturally, although seldom, which demonstrate the importance of p53 methylation. Mutation of Arg337 (normally mutated to cysteine or histidine), related with tumor development, changes biochemical characteristics of p53 (53) and causes tetramer of p53 dynamically unstable (54). Thus, the role of p53 arginine methylation in the regulation of p53 activity may be a mechanism for the effect of these mutations.
Ubiquitination
In normal non-stimulated cells, p53 renews quickly and the expression of p53 remains low level. MDM2 is an important factor for maintaining p53 levels, which promoted the polyubiquitination of p53 and the degradation by proteasome pathway, thereby inhibiting p53-mediated transactivation (55). The major ubiquitination sites of p53 mediated by MDM2 are six lysines at the carboxy terminus (K370, K372, K373, K381, K382, and K386) (56). The expression of MDM2 is also regulated by p53, indicating a negative feedback of p53 expression. So that an increase in p53 level can induce MDM2 expression, leading to a decrease in p53 expression and activity (57). Moreover, several lysines of the DNA binding domain of p53 are also target for ubiquitination (58). MDM4 is similar to MDM2 and inhibits p53-mediated transactivation. The induction of p53 can result from its release from these negative regulatory factors. Inhibits and/or rapidly degradations of MDM2 and MDM4 cause rapid accumulation of p53 and activate its transcription function (59,60). Ubiquitination prevents p53 from binding to the downstream gene in the nucleus, leading to apoptosis and cell cycle arrest (61,62).
A number of E3 ligases involved in MDM2-independent p53 ubiquitination have been identified, such as Pirh2, ICP0, COP1, TOPORS, ARF-BP1, CHIP, Ubc13, synoviolin, EF41, CARP2, WWP1, MSL2, E6-AP, TRIM2454 and MKRN1 (63). Ubc13, WWP1, E4F1 and MSL2 are E3 ligases mediating proteasome-independent ubiquitination of p53. Besides these E3 ligases, MDM2 at low level also mediates mono-ubiquitination of p53, resulting in proteasome-independent p53 ubiquitination (64).
Different types of p53 ubiquitination result in different effects of p53 function. Several E3 ligases, in addition to MDM2, can mediate K48-linked polyubiquitination of p53 and target it to the 26S proteasome for degradation. Other types of ubiquitination, including mono- or K63-linked polyubiquitinations, affect p53 stabilization by regulating nuclear export and cytosolic localizations of p53. Ubiquitination also disrupts p53 from binding to the promoter of target genes as a transcription factor in the nucleus that results in apoptosis and cell cycle arrest (65).
Other post-translational modification
In addition to the modifications described above, there are some other types of modifications of p53 which have already been identified, such as SUMO-1 and SUMO-2/3-mediated sumoylation (66), neddylation (NEDD8) (67), etc. Moreover, O-linked N-acetylglucosamine, ADP-ribosylation and prolyl isomerization modifications can also regulate p53 activity (68). Recently, p53 β-hydroxybutyrylation, a new PTM, has been identified by Wenhui Zhao in 2019, which is catalyzed by CBP and results in lower levels of p53 acetylation, thereby attenuates p53 activity (69).
We have summarized the various PTMs of p53 and their effect on cellular functions (Table 1).
Table 1
| PTM type | PTM residues | Molecular and cellular consequences | Refs. |
|---|---|---|---|
| Phosphorylation | S6, S9 | Regulation of tumorigenesis and metastasis; involves in mesodermal development | ( |
| S15 | Mediates cell cycle G1/S block; increases stabilization of p53 protein in cell stress response | ( |
|
| S37 | Promotes p53 activity in cell stress response | ( |
|
| S46, T55 | Exert apoptotic cell death | ( |
|
| S392 | Activates the sequence-specific DNA binding capacity of p53 | ( |
|
| Acetylation | K305, K370, K372, K386 | Increase the sequence-specific DNA binding capacity | ( |
| K120 | Promotes p53-dependent apoptosis induced by DNA damage; regulates tumor development | ( |
|
| K164 | Regulation of p53-mediated cell growth arrest; promotes p53-dependent apoptosis induced by DNA damage | ( |
|
| K320 | Increases the sequence-specific DNA binding capacity, promotes the recruitment of transcriptional activators, affects p53 shuttle between nucleus and cytoplasm and the p53-dependent BAX-mediated apoptosis following DNA damage; induces cell cycle arrest | ( |
|
| K373, K382 | Promote its promoter-specific transactivity and apoptosis in the cellular UVB response; mediate p21 activation for G1 phase arrest; Deacetylation of K382 is catalyzed by HDAC1 and SIRT1 | ( |
|
| K381 | Activates transcription and induces apoptosis | ( |
|
| Methylation | K370 | Represses the transactivation of p53, resulting in inhibition of p53-mediated cell cycle arrest and apoptosis; Demethylation of K370 is catalyzed by KDM1, prevents the binding of p53 coactivator 53BP1 | ( |
| K370me2 | Positive regulation of p53 activity | ( |
|
| K372 | Enhances the overall stability of p53 binding to chromatin, increases recruitment of p53 in regulatory regions of p21 and other downstream genes | ( |
|
| K373me2 | Inactivation of p53 | ( |
|
| K382 | Reduces the transactivation of p53 to high response downstream genes | ( |
|
| K382me2 | Increases the transactivation of p53 | ( |
|
| R333, R335me2 | Regulation of sequence-specific DNA binding capacity and oligomerization of p53 | ( |
|
| R337me2 | Modulates the promoter binding specificity of p53, changes biochemical characteristics of p53 and causes tetramer of p53 dynamically unstable; regulates tumor development | ( |
|
| Ubiquitination | K370, K372, K373, K381, K382, K386 | Mediates degradation of p53, inhibits p53-mediated transactivation leading to apoptosis and cell cycle arrest | ( |
| Sumoylation | K386 | Represses transcription activity and binding to the endogenous p21 gene of p53 | ( |
| β-hydroxybutyrylation | K120, K319, K370 | Results in lower levels of p53 acetylation, thereby attenuates p53 activity | ( |
PTM, post-transcriptional modification.
Crosstalk between p53 post-translational modifications
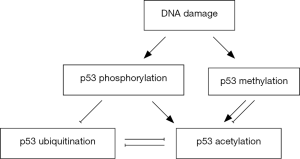
Hupp et al. proposes an allosteric model depending on post-translational modifications of the p53 C-terminus that activates p53 function as a DNA-binding protein, that the C-terminal of p53 can act as a negative regulator, maybe by binding to the core DNA binding region of p53 and make it form an inactive conformation (74). A variety of studies have supported this allosteric model (75,76). Some alterations, such as post-translational modifications and binding of single-strand DNA or antibody can disrupt the interaction between the C-terminal domain and the core domain of p53, allowing the DNA binding domain to form an activated conformation (23,75,76). Importantly, there is close interaction between post-translational modifications of p53 and these modifications participate in the regulation of p53 activity cooperatively.
Activation of p53 triggered by cell stress response is primarily regulated by post-translational modifications of p53, including phosphorylation and acetylation (77). For example, in the UV or radiation-induced response, the first modification induced in the N-terminus of p53 is phosphorylation at Ser33 and Ser37 and then phosphorylated p53 promotes acetylation at K373/K382 and K320 mediated by p300 and PCAF, respectively (78). Phosphorylation of p53 at the C-terminus induced by CHK1 and CHK2 in DNA damage response also regulates acetylation of the C-terminus (79). In the DNA damage response, both acetylation at K392 and phosphorylation at Ser392 of p53 increase the interaction between p53 and MDC1, which is an important linker protein capable of recruiting many proteins to DNA damage sites (80). Phosphorylation of the p53 C-terminus regulatory region (such as Ser392) catalyzed by casein kinase II (CK2) promotes binding of p53 with DNA and induces site-specific acetylation dependent on DNA and p300 (75). Phosphorylation of p53 amino terminal sites, including Ser15, Ser20, Ser33, Ser37, Ser46, Ser55 and Thr18 promotes the binding of p53 with p300/CBP and the transactivation of p53 (81). In addition, the bisphosphorylation or polyphosphorylation events cooperatively increase the interaction of p53 and p300 by about 80-fold (82). Phosphorylation at these sites also prevents MDM2 binding, resulting in a decrease in p53 renewal (83).
Ubiquitination and acetylation are mutually exclusive events that have different effect on the regulation of p53 functions. With ubiquitination mediated by MDM2, p53 cannot be acetylated by p300/CBP, leading to rapid degradation through proteasome pathway (84). Interestingly, p300/CBP not only acetylated p53, but also acetylated MDM2, resulting in inhibition of p53 ubiquitination mediated by MDM2 (85). Acetylation of p53 inhibits its interaction with MDM2 and MDM4 (24,68). Recent studies have demonstrated that tripartite motif-containing protein 25 (TRIM25) might be a negative regulator of p53 acetylation and polyubiquitination as well as a positive modulator for p53 sumoylation, which modulates p53 nuclear export in prostate cancer cells (86).
There is also close relationship between methylation and acetylation at p53 lysine residues. Lysine methylation modification mainly occurs in response to DNA damage, which will promote (44,87) or even inhibit (48) the subsequent acetylation at other residues. For example, methylation at p53 K372 has the function to induce the subsequent acetylation modifications, thereby increase the stability and activity of p53 and result in upregulation of its target p21 gene and cell cycle arrest. p53 acetylation induced by DNA damage is also impaired in the absence of lysine methylation (87). Taken together, there is close and intricate relationship among post-translational modifications of p53, which is crucial for the function of p53 in cell stress and tumor suppression. Figure 1 has exhibited the crosstalk among post-translational modifications of p53 in response of DNA damage.
Summary and outlook
Different stimuli lead to different post-translational modifications of p53, making p53 a protein that can be dynamically regulated, and play the corresponding function quickly and accurately in various stress responses and physiological processes. As the most frequently inactivated tumor suppressor in human cancers, the inactivation and mutation of p53 has been reported in more than 50% of cancers (88). It has been widely accepted that regulation on cell cycle progression is one of the major mechanisms by which p53 inhibits tumor cell growth (89). As an important transcription factor, p53 facilitates cell cycle arrest mainly through upregulation of the expression of its target genes involved in cell cycle progression, such as p21, GADD45 and Cdc25C (90). Since PTMs of p53 play essential roles in regulation transactivity of p53, cell cycle progression mediated by p53 is also regulated by these modifications. We have summarized the effect of various p53 PTMs on cell cycle progression (Table 2). Interaction between post-translational modifications makes the regulation of p53 function complex and delicate. All of the post-translational modifications of p53 are reversible. PTM-removal enzymes are important for cells to recover from stress response and play an essential role in establishing thresholds for p53 activation, thereby preventing p53 from improper activation. Furthermore, some modifications are mutually exclusive, so PTM-removal enzymes are required to change the state of modification, in particular for the activation of p53 to promote its function. These PTM-removal enzymes can affect the development of tumors, which may serve as a potential target for new antineoplastic drugs. Considering that p53 also inhibits the expression of many genes, the role of post-translational modifications in p53-mediated transcriptional inhibition is also an interesting subject to research. Moreover, since p53 is an important tumor suppressor and is dysfunctional in a variety of cancers, does the post-translational modification, in addition to mutations in gene sequences, lead to abnormal activity of p53 in these cancers? Research on the function of p53 post-translational modifications, its underlying molecular mechanism(s) and interaction between these modifications are of great significance to provide new molecular basis for therapeutic strategy of p53 dysfunction associated cancers.
Table 2
| PTM type | Effect on cell cycle progression | Refs. |
|---|---|---|
| Phosphorylation | ATM and ATR phosphorylate p53 (Ser6, -15, -37, and -392) in response to DNA damage mediate p21 activation for arresting the cell cycle at the G1-S checkpoint | ( |
| Phosphorylation of p53 on Ser15 induced by AMPK activation initiates AMPK-dependent cell-cycle arrest (G1/S block) | ( |
|
| Phosphorylation of p53 on Ser33 mediated by p38γ, p38δ and JNK2 causes a transient G1 arrest | ( |
|
| Acetylation | Acetylation of p53 at K164, K320, K373 and K382 induce cell cycle arrest | ( |
| Methylation | Methylation of p53 at R333, R335 and R337 enhance p53-dependent cell cycle arrest | ( |
| Smyd2-mediated monomethylation at p53K370 inhibits p53-mediated cell cycle arrest | ( |
|
| Methylation of p53K372 induces cell cycle arrest by upregulation of p53 target p21 gene | ( |
|
| Ubiquitination | E3 ligase Pirh2 mediated ubiquitination of p53 decreases p53-mediated cell cycle arrest | ( |
| K48-linked ubiquitination of p53 mediated by E3 ligase MDM2 inhibits cell cycle arrest | ( |
PTM, post-transcriptional modification.
Acknowledgments
Funding: This work was supported by a grant from the Natural Science Foundation of Shandong Province (No. ZR2018BC019).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://fomm.amegroups.org/article/view/10.21037/fomm.2019.12.02/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Malkin D, Li FP, Strong LC, et al. Germ Line P53 Mutations in a Familial Syndrome of Breast-Cancer, Sarcomas, and Other Neoplasms. Science 1990;250:1233-8. [Crossref] [PubMed]
- Donehower LA, Harvey M, Slagle BL, et al. Mice Deficient for P53 Are Developmentally Normal but Susceptible to Spontaneous Tumors. Nature 1992;356:215-21. [Crossref] [PubMed]
- Hollstein M, Sidransky D, Vogelstein B, et al. P53 Mutations in Human Cancers. Science 1991;253:49-53. [Crossref] [PubMed]
- Bennett WP, Hollstein MC, Hsu IC, et al. Mutational spectra and immunohistochemical analyses of p53 in human cancers. Chest 1992;101:19S-20S. [Crossref] [PubMed]
- el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol 1998;8:345-57. [Crossref] [PubMed]
- Zhao R, Gish K, Murphy M, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev 2000;14:981-93. [PubMed]
- Brady CA, Attardi LD. p53 at a glance. J Cell Sci 2010;123:2527-32. [Crossref] [PubMed]
- Morgan SE, Kastan MB. p53 and ATM: cell cycle, cell death, and cancer. Adv Cancer Res 1997;71:1-25. [Crossref] [PubMed]
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 2001;268:2764-72. [Crossref] [PubMed]
- Olsson A, Manzl C, Strasser A, et al. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ 2007;14:1561-75. [Crossref] [PubMed]
- Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol 2009;1:a000950 [Crossref] [PubMed]
- Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med 2010;16:528-36. [Crossref] [PubMed]
- Samad A, Anderson CW, Carroll RB. Mapping of Phosphomonoester and Apparent Phosphodiester Bonds of the Oncogene Product P53 from Simian-Virus 40-Transformed 3t3 Cells. Proc Natl Acad Sci U S A 1986;83:897-901. [Crossref] [PubMed]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, et al. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol 1992;12:5041-9. [Crossref] [PubMed]
- Shieh SY, Ikeda M, Taya Y, et al. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997;91:325-34. [Crossref] [PubMed]
- Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular Cell 2005;18:283-93. [Crossref] [PubMed]
- Adorno M, Cordenonsi M, Montagner M, et al. A Mutant-p53/Smad Complex Opposes p63 to Empower TGF beta-Induced Metastasis. Cell 2009;137:87-98. [Crossref] [PubMed]
- Cordenonsi M, Montagner M, Adorno M, et al. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science 2007;315:840-3. [Crossref] [PubMed]
- Mancini F, Pieroni L, Monteleone V, et al. MDM4/HIPK2/p53 cytoplasmic assembly uncovers coordinated repression of molecules with anti-apoptotic activity during early DNA damage response. Oncogene 2016;35:228-40. [Crossref] [PubMed]
- Taira N, Yamaguchi T, Kimura J, et al. Induction of amphiregulin by p53 promotes apoptosis via control of microRNA biogenesis in response to DNA damage. Proc Natl Acad Sci U S A 2014;111:717-22. [Crossref] [PubMed]
- Di Lello P, Jenkins LM, Jones TN, et al. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell 2006;22:731-40. [Crossref] [PubMed]
- Chène P. The role of tetramerization in p53 function. Oncogene 2001;20:2611-7. [Crossref] [PubMed]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997;90:595-606. [Crossref] [PubMed]
- Tang Y, Zhao WH, Chen Y, et al. Acetylation is indispensable for p53 activation. Cell 2008;133:612-26. [Crossref] [PubMed]
- Liu Y, Colosimo AL, Yang XJ, et al. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol Cell Biol 2000;20:5540-53. [Crossref] [PubMed]
- Barlev NA, Liu L, Chehab NH, et al. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 2001;8:1243-54. [Crossref] [PubMed]
- Basile V, Mantovani R, Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J Biol Chem 2006;281:2347-57. [Crossref] [PubMed]
- Imbriano C, Gurtner A, Cocchiarella F, et al. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol 2005;25:3737-51. [Crossref] [PubMed]
- Feng L, Lin T, Uranishi H, et al. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol 2005;25:5389-95. [Crossref] [PubMed]
- Krummel KA, Lee CJ, Toledo F, et al. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A 2005;102:10188-93. [Crossref] [PubMed]
- Ito A, Kawaguchi Y, Lai CH, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 2002;21:6236-45. [Crossref] [PubMed]
- Glozak MA, Sengupta N, Zhang XH, et al. Acetylation and deacetylation of non-histone proteins. Gene 2005;363:15-23. [Crossref] [PubMed]
- Smith J. Human Sir2 and the 'silencing' of p53 activity. Trends Cell Biol 2002;12:404-6. [Crossref] [PubMed]
- Luo J, Li M, Tang Y, et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A 2004;101:2259-64. [Crossref] [PubMed]
- Wang YH, Tsay YG, Tan BC, et al. Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J Biol Chem 2003;278:25568-76. [Crossref] [PubMed]
- Berns K, Hijmans EM, Mullenders J, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 2004;428:431-7. [Crossref] [PubMed]
- Arbely E, Natan E, Brandt T, et al. Acetylation of lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc Natl Acad Sci U S A 2011;108:8251-6. [Crossref] [PubMed]
- Joubel A, Chalkley RJ, Medzihradszky KF, et al. Identification of New p53 Acetylation Sites in COS-1 Cells. Mol Cell Proteomics 2009;8:1167-73. [Crossref] [PubMed]
- Kusio-Kobialka M, Wolanin K, Podszywalow-Bartnicka P, et al. Increased acetylation of lysine 317/320 of p53 caused by BCR-ABL protects from cytoplasmic translocation of p53 and mitochondria-dependent apoptosis in response to DNA damage. Apoptosis 2012;17:950-63. [Crossref] [PubMed]
- Li L, Jing L, Wang J, et al. Autophagic flux is essential for the downregulation of D-dopachrome tautomerase by atractylenolide I to ameliorate intestinal adenoma formation. J Cell Commun Signal 2018;12:689-98. [Crossref] [PubMed]
- Kern SE, Kinzler KW, Bruskin A, et al. Identification of p53 as a sequence-specific DNA-binding protein. Science 1991;252:1708-11. [Crossref] [PubMed]
- Itahana Y, Ke H, Zhang Y. p53 Oligomerization is essential for its C-terminal lysine acetylation. J Biol Chem 2009;284:5158-64. [Crossref] [PubMed]
- Huang J, Sengupta R, Espejo AB, et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007;449:105-8. [Crossref] [PubMed]
- Kurash JK, Lei H, Shen Q, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell 2008;29:392-400. [Crossref] [PubMed]
- Shi X, Kachirskaia I, Yamaguchi H, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 2007;27:636-46. [Crossref] [PubMed]
- Jansson M, Durant ST, Cho EC, et al. Arginine methylation regulates the p53 response. Nat Cell Biol 2008;10:1431-9. [Crossref] [PubMed]
- Chuikov S, Kurash JK, Wilson JR, et al. Regulation of p53 activity through lysine methylation. Nature 2004;432:353-60. [Crossref] [PubMed]
- Huang J, Perez-Burgos L, Placek BJ, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006;444:629-32. [Crossref] [PubMed]
- Kachirskaia I, Shi XB, Yamaguchi H, et al. Role for 53BP1 Tudor Domain Recognition of p53 Dimethylated at Lysine 382 in DNA Damage Signaling. J Biol Chem 2008;283:34660-6. [Crossref] [PubMed]
- Huang J, Dorsey J, Chuikov S, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 2010;285:9636-41. [Crossref] [PubMed]
- Bedford MT, Richard S. Arginine methylation: An emerging regulator of protein function. Molecular Cell 2005;18:263-72. [Crossref] [PubMed]
- Vousden KH. p53 and PUMA: A deadly duo. Science 2005;309:1685-6. [Crossref] [PubMed]
- Lomax ME, Barnes DM, Hupp TR, et al. Characterization of p53 oligomerization domain mutations isolated from Li-Fraumeni and Li-Fraumeni like family members. Oncogene 1998;17:643-9. [Crossref] [PubMed]
- Kawaguchi T, Kato S, Otsuka K, et al. The relationship among p53 oligomer formation, structure and transcriptional activity using a comprehensive missense mutation library. Oncogene 2005;24:6976-81. [Crossref] [PubMed]
- Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [Crossref] [PubMed]
- Rodriguez MS, Desterro JM, Lain S, et al. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol 2000;20:8458-67. [Crossref] [PubMed]
- Wu X, Bayle JH, Olson D, et al. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993;7:1126-32. [Crossref] [PubMed]
- Chan WM, Mak MC, Fung TK, et al. Ubiquitination of p53 at multiple sites in the DNA-binding domain. Mol Cancer Res 2006;4:15-25. [Crossref] [PubMed]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006;6:909-23. [Crossref] [PubMed]
- Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 2009;9:714-23. [Crossref] [PubMed]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J 2005;24:3353-9. [Crossref] [PubMed]
- Lee JT, Wheeler TC, Li L, et al. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet 2008;17:906-17. [Crossref] [PubMed]
- Lee EW, Lee MS, Camus S, et al. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J 2009;28:2100-13. [Crossref] [PubMed]
- Li MY, Brooks CL, Wu-Baer F, et al. Mono-versus polyubiquitination: Differential control of p53 fate by Mdm2. Science 2003;302:1972-5. [Crossref] [PubMed]
- Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 2010;17:86-92. [Crossref] [PubMed]
- Bischof O, Schwamborn K, Martin N, et al. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol Cell 2006;22:783-94. [Crossref] [PubMed]
- Xirodimas DP, Saville MK, Bourdon JC, et al. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 2004;118:83-97. [Crossref] [PubMed]
- Kruse JP, Gu W. Modes of p53 Regulation. Cell 2009;137:609-22. [Crossref] [PubMed]
- Liu K, Li F, Sun Q, et al. p53 beta-hydroxybutyrylation attenuates p53 activity. Cell Death Dis 2019;10:243. [Crossref] [PubMed]
- Koh DI, Han D, Ryu H, et al. KAISO, a critical regulator of p53-mediated transcription of CDKN1A and apoptotic genes. Proc Natl Acad Sci U S A 2014;111:15078-83. [Crossref] [PubMed]
- Bao L, Diao H, Dong N, et al. Histone deacetylase inhibitor induces cell apoptosis and cycle arrest in lung cancer cells via mitochondrial injury and p53 up-acetylation. Cell Biol Toxicol 2016;32:469-82. [Crossref] [PubMed]
- Brochier C, Dennis G, Rivieccio MA, et al. Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J Neurosci 2013;33:8621-32. [Crossref] [PubMed]
- Wu SY, Chiang CM. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J 2009;28:1246-59. [Crossref] [PubMed]
- Hupp TR, Meek DM, Midgley CA, et al. Regulation of the specific DNA binding function of p53. Cell 1992;71:875-86. [Crossref] [PubMed]
- Hupp TR, Sparks A, Lane DP. Small Peptides Activate the Latent Sequence-Specific DNA-Binding Function of P53. Cell 1995;83:237-45. [Crossref] [PubMed]
- Jayaraman J, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 1995;81:1021-9. [Crossref] [PubMed]
- Appella E, Anderson CW. Signaling to p53: breaking the posttranslational modification code. Pathol Biol (Paris) 2000;48:227-45. [PubMed]
- Sakaguchi K, Herrera JE, Saito S, et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 1998;12:2831-41. [Crossref] [PubMed]
- Ou YH, Chung PH, Sun TP, et al. P53C-terminal phosphorylation by CHM and CHK2 participates in the regulation of DNA-Damage-induced C-terminal acetylation. Mol Biol Cell 2005;16:1684-95. [Crossref] [PubMed]
- Shahar OD, Gabizon R, Feine O, et al. acetylation of lysine 382 and phosphorylation of serine 392 in p53 modulate the interaction between p53 and MDC1 in vitro. PLoS One 2013;8:e78472 [Crossref] [PubMed]
- Feng H, Jenkins LM, Durell SR, et al. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure 2009;17:202-10. [Crossref] [PubMed]
- Teufel DP, Bycroft M, Fersht AR. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene 2009;28:2112-8. [Crossref] [PubMed]
- Ferreon JC, Lee CW, Arai M, et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci U S A 2009;106:6591-6. [Crossref] [PubMed]
- Ito A, Lai CH, Zhao X, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J 2001;20:1331-40. [Crossref] [PubMed]
- Wang X, Taplick J, Geva N, et al. Inhibition of p53 degradation by Mdm2 acetylation. FEBS Lett 2004;561:195-201. [Crossref] [PubMed]
- Takayama KI, Suzuki T, Tanaka T, et al. TRIM25 enhances cell growth and cell survival by modulating p53 signals via interaction with G3BP2 in prostate cancer. Oncogene 2018;37:2165-80. [Crossref] [PubMed]
- Ivanov GS, Ivanova T, Kurash J, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 2007;27:6756-69. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Li T, Kon N, Jiang L, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012;149:1269-83. [Crossref] [PubMed]
- Peng CY, Graves PR, Thoma RS, et al. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 1997;277:1501-5. [Crossref] [PubMed]
- Siliciano JD, Canman CE, Taya Y, et al. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev 1997;11:3471-81. [Crossref] [PubMed]
- Hawkes WC, Alkan Z. Delayed cell cycle progression in selenoprotein W-depleted cells is regulated by a mitogen-activated protein kinase kinase 4-p38/c-Jun NH2-terminal kinase-p53 pathway. J Biol Chem 2012;287:27371-9. [Crossref] [PubMed]
- Basbous J, Knani D, Bonneaud N, et al. Induction of ASAP (MAP9) contributes to p53 stabilization in response to DNA damage. Cell Cycle 2012;11:2380-90. [Crossref] [PubMed]
- Cui D, Li L, Lou H, et al. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene 2014;33:2225-35. [Crossref] [PubMed]
- Reed SM, Quelle DE. p53 Acetylation: Regulation and Consequences. Cancers (Basel) 2014;7:30-69. [Crossref] [PubMed]
- Raposo AE, Piller SC. Protein arginine methylation: an emerging regulator of the cell cycle. Cell Div 2018;13:3. [Crossref] [PubMed]
- Leng RP, Lin Y, Ma W, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003;112:779-91. [Crossref] [PubMed]
- Jin J, Lin G, Huang H, et al. Capsaicin mediates cell cycle arrest and apoptosis in human colon cancer cells via stabilizing and activating p53. Int J Biol Sci 2014;10:285-95. [Crossref] [PubMed]
- Li L, Li W, Xiao L, et al. Viral oncoprotein LMP1 disrupts p53-induced cell cycle arrest and apoptosis through modulating K63-linked ubiquitination of p53. Cell Cycle 2012;11:2327-36. [Crossref] [PubMed]
Cite this article as: Che Z, Sun H, Yao W, Lu B, Han Q. Role of post-translational modifications in regulation of tumor suppressor p53 function. Front Oral Maxillofac Med 2020;2:1.